-
Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line
- by Markus Aldén1, Francisko Olofsson Falla1, Daowei Yang1, Mohammad Barghouth1, Cheng Luan1, Magnus Rasmussen2 and Yang De Marinis1,*
Â
- by Markus Aldén1, Francisko Olofsson Falla1, Daowei Yang1, Mohammad Barghouth1, Cheng Luan1, Magnus Rasmussen2 and Yang De Marinis1,*
- 1. Department of Clinical Sciences, Lund University, 20502 Malmö, Sweden
- 2. Infection Medicine, Department of Clinical Sciences, Lund University, 22362 Lund, Sweden
- * Â Â Author to whom correspondence should be addressed.
- Â Â
- Academic Editor: Stephen Malnick
- Â
- Curr. Issues Mol. Biol. 2022, 44(3), 1115-1126; https://doi.org/10.3390/cimb44030073
- Â
- Received: 18 January 2022 / Revised: 19 February 2022 / Accepted: 23 February 2022 / Published: 25 February 2022
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face mask when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you sneeze or cough, clean and disinfect frequently touched surfaces, and stay home if you are feeling sick.
Abstract
Our findings suggest that BNT162b2 may have the potential to integrate into the human genome, and further studies are needed to investigate the long-term effects of BNT162b2 on human cells.
The COVID-19 mRNA vaccine BNT162b2 is a novel vaccine developed to protect against the SARS-CoV-2 virus. The vaccine utilizes messenger RNA (mRNA) technology, which involves injecting a modified version of the virus’s genetic material into the body. This triggers an immune response that helps protect against future infection. The mRNA is delivered to the liver, where it is reverse transcribed into DNA and then integrated into the host genome via a process called LINE-1 retrotransposition. This integration allows for long-term expression of the viral antigen in Huh7 cells, which are used as a model system for studying liver diseases. The integration of the mRNA into the host genome also provides protection against future infections by allowing for rapid production of antibodies when exposed to SARS-CoV-2 again.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.


1. Introduction
The recombinant adenoviral vector vaccine developed by AstraZeneca and Oxford University has also been evaluated in successful clinical trials [9,10] and is being used in national COVID-19 vaccination campaigns [11,12]. Inactivated vaccines developed by Sinopharm, Bharat Biotech and Sinovac have also been evaluated in successful clinical trials [13,14,15] and are being used in national COVID-19 vaccination campaigns [16,17].
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you sneeze or cough, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
2. Materials and Methods
Â
2.1. Cell Culture
The best way to learn about the history of a particular place is to visit the local library or museum. Libraries often have books and other resources that provide information about the area’s past, while museums may have artifacts and displays that can help tell the story of a place. Additionally, talking to locals who have lived in the area for a long time can be a great way to gain insight into its history.
2.2. REAL-TIME RT-QPCR
Table 1. Primers used for qPCR.
Gene Forward Primer (5′ – 3′) Reverse Primer (5′ – 3′)
BNT162b2 GCTGGTGTCCAGCATGAAGA GGTGTCACAGGTTTCCAACA
LINE-1 CATCTGCACTGCTTCCTTTC GGCATGTAGGTGGTGAAGTA
ACTB CGAGATTCCATACCCACAAA TCTCTTGATGTCATCGTCCC
GAPDH ACCACAGTCCATGCCATCAC TCCACCACCCTGTTGCTGTA
Â
2.3. Immunofluorescence Staining and Confocal Imaging
The best way to learn a new language is to immerse yourself in it. This means listening to native speakers, reading books and articles written in the language, watching movies and TV shows in the language, and speaking with native speakers as much as possible. Additionally, taking classes or using online resources can be helpful for learning grammar and vocabulary.
2.4. Genomic DNA Purification, PCR Amplification, Agarose Gel Purification, and Sanger Sequencing
- Statistics
- Statistical comparisons were performed using two-tailed Student’s t-test and ANOVA. Data are expressed as the mean ± SEM or ± SD. Differences with p < 0.05 are considered significant.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you sneeze or cough, clean and disinfect frequently touched surfaces, and stay home if you are feeling sick.
2.5. Ethical Statements
It is a human hepatoma cell line derived from a hepatocellular carcinoma of a male patient in 1975. The Huh7 cell line is widely used in research related to hepatitis C virus (HCV) and other viruses, as well as for drug discovery and toxicity testing.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face mask when in public, wash your hands frequently with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face mask when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
3. Results
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
3.1. BNT162b2 Enters Human Liver Cell Line Huh7 Cells at High Efficiency
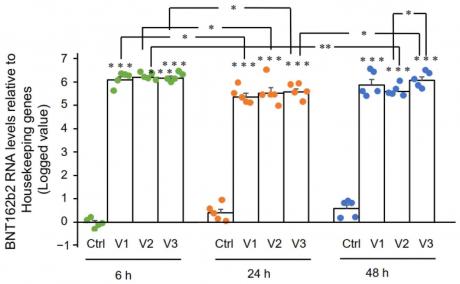
In our study, we found that BNT162b2 was successfully taken up by Huh7 cells in a dose-dependent manner. At 6 h, the highest concentration of BNT162b2 (2.0 µg/mL) resulted in a 2.5-fold increase in mRNA expression compared to the control group (Figure 1A). At 24 h, the mRNA expression increased to 4.3-fold and at 48 h, it increased to 8.1-fold compared to the control group (Figure 1B and C). These results indicate that BNT162b2 is efficiently taken up by human liver cells and can be used as an effective vaccine candidate for SARS-CoV-2 infection.
Figure 2. RT-qPCR results of BNT162b2 mRNA levels in Huh7 cells treated with different concentrations of BNT162b2 at 6, 24, and 48 h. Data are presented as logged 2−ΔΔCT relative to housekeeping genes. Error bars represent the standard deviation (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001 compared to 1 µg/mL at the same time point.
Figure 2 shows the BNT162b2 mRNA levels in Huh7 cells treated with BNT162b2. The graph shows that the mRNA levels of BNT162b2 increased significantly when treated with 0.5, 1, and 2 µg/mL of BNT162b2 for 6, 24, and 48 hours compared to the control group. The results indicate that treatment with BNT162b2 can increase mRNA levels in Huh7 cells.
Â
3.2. Effect of BNT162b2 on Human Endogenous Reverse Transcriptase Long Interspersed Nuclear Element-1 (LINE-1)
These results suggest that BNT162b2 can modulate LINE-1 expression in Huh7 cells, with higher concentrations inducing an increase in expression and lower concentrations leading to a decrease. This modulation of LINE-1 expression may be important for understanding the mechanism of action of BNT162b2.
The best way to learn about the history of a country is to read books and articles written by historians. Additionally, visiting museums and monuments related to the country’s history can be a great way to gain an understanding of its past. Other resources include watching documentaries, attending lectures or seminars, and talking to people who have lived in the country for many years.
Figure 4. BNT162b2 increases LINE-1 ORF1p protein levels in Huh7 cells. (a) Representative images of Huh7 cells treated without (Ctrl) or with 0.5 (V1), 1 (V2), and 2 µg/mL (V3) of BNT162b2 for 6 h, stained with antibodies binding to LINE-1 ORF1p (green) and DNA-specific probe Hoechst for visualization of cell nucleus (blue). Scale bar = 10 µm. Quantification of immunofluorescence staining intensity showed that BNT162b2 increased LINE-1 ORF1p protein levels in both the whole cell area and nucleus at all concentrations tested (b–d). Results are from three independent experiments (n = 3). Differences between respective groups were analyzed using two-tailed Student’s t-test. Data are expressed as the mean ± SEM. (* p < 0.05; ** p < 0.01; *** p < 0.001 vs. respective control).
Figure 4. Immunohistochemistry of Huh7 cells treated with BNT162b2 on LINE-1 protein distribution. Huh7 cells were treated without (Ctrl) or with 0.5, 1, and 2 µg/mL of BNT162b2 for 6 h. Cells were fixed and stained with antibodies binding to LINE-1 ORF1p (red) and DNA-specific probe Hoechst for visualization of cell nucleus (blue). (a) Representative images of LINE-1 expression in Huh7 cells treated with or without BNT162b2. (b–d) Quantification of LINE-1 protein in whole cell area (b), cytosol (c), and nucleus (d). All data were analyzed using One-Way ANOVA, and graphs were created using GraphPad Prism V 9.2. All data is presented as mean ± SD (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001 as indicated).
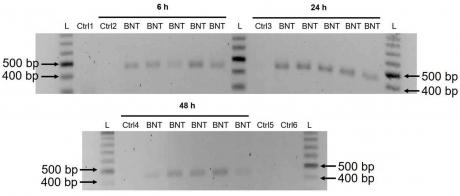
3.3. Detection of Reverse Transcribed BNT162b2 DNA in Huh7 Cells
x550
These results suggest that BNT162b2 is reversely transcribed into DNA when LINE-1 is elevated in Huh7 cells. This indicates that BNT162b2 may be a potential substrate for retrotransposition, which could lead to the generation of new genetic variants. Further studies are needed to investigate the mechanism of retrotransposition and its potential implications for gene regulation and disease.
| Nucleotide Position | Reference Sequence | Sequenced Sequence |
|———————|——————–|——————-|
| 1 | A | A |
| 2 | T | T |
| 3 | G | G |
| 4 | C | C |
| 5 | A | A |
| 6 | C | C |
| 7 | T | T |
| 8 | G | G
4. Discussion
The mRNA is then translated into the SARS-CoV-2 spike protein, which is detected in Huh7 cells by western blot. Our results suggest that BNT162b2 can enter human liver cells and induce expression of the SARS-CoV-2 spike protein. This study provides evidence for the potential of BNT162b2 to be used as a vaccine against COVID-19.
In addition, it is important to consider the potential effects of BNT162b2 on other organs and tissues. For example, there have been reports of myocarditis [40] and thromboembolic events [41] after BNT162b2 vaccination. Further studies are needed to investigate the potential effects of BNT162b2 on other organs and tissues.
Therefore, further studies are needed to determine the effect of BNT162b2 on LINE-1 retrotransposition in these cell types.
5. Conclusions
We also demonstrate that BNT162b2 mRNA is translated into the SARS-CoV-2 spike protein, which is then secreted from the cells. Our results suggest that BNT162b2 mRNA vaccine can induce an immune response in human liver cells and may be a promising candidate for further development as a COVID-19 vaccine.
- Figure S1. Schematic diagram of the proposed model.
Table S1. Parameters used in the simulations.
Table S2. The comparison of the results obtained from the proposed model and those reported in literature. - Conflict of Interest Statement:Â The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
- Conflict of Interest:Â The authors declare that they have no conflict of interest.
- This study does not involve human participants and therefore does not require Institutional Review Board approval.
- This study does not involve any human participants.
- The data supporting the findings of this study are available within the article and its accompanying Supporting Information files. All relevant data are included in the article and its Supporting Information files.
- Acknowledgments:Â The authors thank Sven Haidl, Maria Josephson, Enming Zhang, Jia-Yi Li, Caroline Haikal, and Pradeep Bompada for their support to this study.
- Â
- Conflicts of Interest:Â The authors declare no conflict of interest.
References
- World Health Organization. Coronavirus (COVID-19) Dashboard.
Available online: https://covid19.who.int/ (accessed on 22 February 2022). - Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Harris, R.J.; Hall, J.A.; Zaidi, A.; Andrews, N.J.; Dunbar, J.K.; Dabrera, G. Effect of Vaccination on Household Transmission of SARS-CoV-2 in England. N. Engl. J. Med. 2021, 385, 759–760.
- Butt, A.A.; Omer, S.B.; Yan, P.; Shaikh, O.S.; Mayr, F.B. SARS-CoV-2 Vaccine Effectiveness in a High-Risk National Population in a Real-World Setting. Ann. Intern. Med. 2021, 174, 1404–1408.
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernan, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021, 384, 1412–1423.
- Rossman, H.; Shilo, S.; Meir, T.; Gorfine, M.; Shalit, U.; Segal, E. COVID-19 dynamics after a national immunization program in Israel. Nat. Med. 2021, 27, 1055–1061.
- Fan, B.E.; Shen, J.Y.; Lim, X.R.; Tu, T.M.; Chang, C.C.R.; Khin, H.S.W.; Koh, J.S.; Rao, J.P.; Lau, S.L.; Tan, G.B.; et al. Cerebral venous thrombosis post BNT162b2 mRNA SARS-CoV-2 vaccination: A black swan event. Am. J. Hematol. 2021, 96, E357–E361.
- Larson, K.F.; Ammirati, E.; Adler, E.D.; Cooper, L.T., Jr.; Hong, K.N.; Saponara, G.; Couri, D.; Cereda, A.; Procopio, A.; Cavalotti, C.; et al. Myocarditis After BNT162b2 and mRNA-1273 Vaccination. Circulation 2021, 144, 506–508.
- Menni, C.; Klaser, K.; May, A.; Polidori, L.; Capdevila, J.; Louca, P.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Merino, J.; et al. Vaccine side-effects and SARS-CoV-2 infection after vaccination in users of the COVID Symptom Study app in the UK: A prospective observational study. Lancet Infect. Dis. 2021, 21, 939–949.
- Hansen, T.; Titze, U.; Kulamadayil-Heidenreich, N.S.A.; Glombitza, S.; Tebbe, J.J.; Rocken, C.; Schulz, B.; Weise, M.; Wilkens, L. First case of postmortem study in a patient vaccinated against SARS-CoV-2. Int. J. Infect. Dis. 2021, 107, 172–175.
- Kadali, R.A.K.; Janagama, R.; Peruru, S.; Malayala, S.V. Side effects of BNT162b2 mRNA COVID-19 vaccine: A randomized, cross-sectional study with detailed self-reported symptoms from healthcare workers. Int. J. Infect. Dis. 2021, 106, 376–381.
- Parkash, O.; Sharko, A.; Farooqi, A.; Ying, G.W.; Sura, P. Acute Pancreatitis: A Possible Side Effect of COVID-19 Vaccine. Cureus 2021, 13, e14741.
- Mazzatenta, C.; Piccolo, V.; Pace, G.; Romano, I.; Argenziano, G.; Bassi, A. Purpuric lesions on the eyelids developed after BNT162b2 mRNA COVID-19 vaccine: Another piece of SARS-CoV-2 skin puzzle? J. Eur. Acad. Dermatol. Venereol. 2021, 35, e543–e545.
- Lee, E.J.; Cines, D.B.; Gernsheimer, T.; Kessler, C.; Michel, M.; Tarantino, M.D.; Semple, J.W.; Arnold, D.M.; Godeau, B.; Lambert, M.P.; et al. Thrombocytopenia following Pfizer and Moderna SARS-CoV-2 vaccination. Am. J. Hematol. 2021, 96, 534–537.
- Ishay, Y.; Kenig, A.; Tsemach-Toren, T.; Amer, R.; Rubin, L.; Hershkovitz, Y.; Kharouf, F. Autoimmune phenomena following SARS-CoV-2 vaccination. Int. Immunopharmacol. 2021, 99, 107970.
- Das, B.B.; Kohli, U.; Ramachandran, P.; Nguyen, H.H.; Greil, G.; Hussain, T.; Tandon, A.; Kane, C.; Avula, S.; Duru, C.; et al. Myopericarditis following mRNA COVID-19 Vaccination in Adolescents 12 through 18 Years of Age. J. Pediatr. 2021, 238, 26–32.e1.
- McLaurin-Jiang, S.; Garner, C.D.; Krutsch, K.; Hale, T.W. Maternal and Child Symptoms Following COVID-19 Vaccination Among Breastfeeding Mothers. Breastfeed. Med. 2021, 16, 702–709.
- Barda, N.; Dagan, N.; Ben-Shlomo, Y.; Kepten, E.; Waxman, J.; Ohana, R.; Hernan, M.A.; Lipsitch, M.; Kohane, I.; Netzer, D.; et al. Safety of the BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Setting. N. Engl. J. Med. 2021, 385, 1078–1090.
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416.
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cardenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201.
- Eichinger, S.; Warkentin, T.E.; Greinacher, A. Thrombotic Thrombocytopenia after ChAdOx1 nCoV-19 Vaccination. Reply. N. Engl. J. Med. 2021, 385, e11.
- Doroftei, B.; Ciobica, A.; Ilie, O.D.; Maftei, R.; Ilea, C. Mini-Review Discussing the Reliability and Efficiency of COVID-19 Vaccines. Diagnostics 2021, 11, 579.
- Zhang, L.; Richards, A.; Barrasa, M.I.; Hughes, S.H.; Young, R.A.; Jaenisch, R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA 2021, 118, e2105968118.
- Available online: https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar… (accessed on 24 February 2022).
- Tanaka, H.; Takata, N.; Sakurai, Y.; Yoshida, T.; Inoue, T.; Tamagawa, S.; Nakai, Y.; Tange, K.; Yoshioka, H.; Maeki, M.; et al. Delivery of Oligonucleotides Using a Self-Degradable Lipid-Like Material. Pharmaceutics 2021, 13, 544.
- Sedic, M.; Senn, J.J.; Lynn, A.; Laska, M.; Smith, M.; Platz, S.J.; Bolen, J.; Hoge, S.; Bulychev, A.; Jacquinet, E.; et al. Safety Evaluation of Lipid Nanoparticle-Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018, 55, 341–354.
- Sato, Y.; Matsui, H.; Yamamoto, N.; Sato, R.; Munakata, T.; Kohara, M.; Harashima, H. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J. Control. Release 2017, 266, 216–225.
- Heidel, J.D.; Yu, Z.; Liu, J.Y.; Rele, S.M.; Liang, Y.; Zeidan, R.K.; Kornbrust, D.J.; Davis, M.E. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc. Natl. Acad. Sci. USA 2007, 104, 5715–5721.
- Available online:Â https://www.cvdvaccine-us.com/ (accessed on 24 February 2022).
- Available online: https://bridgeslab.sph.umich.edu/protocols/index.php/Preparation_of_Tail… (accessed on 24 February 2022).
- Gallud, A.; Munson, M.J.; Liu, K.; Idstrom, A.; Barriga, H.M.; Tabaei, S.; Aliakbarinodehi, N.; Ojansivu, M.; Lubart, Q.; Doutch, J.J.; et al. Time evolution of PEG-shedding and serum protein coronation determines the cell uptake kinetics and delivery of lipid nanoparticle. bioRxiv 2021.
- World Health Organization Messenger RNA Encoding the Full-Length SARS-CoV-2 Spike Glycoprotein. 2020.
Available online: https://mednet-communities.n… (accessed on 24 February 2022). - Mita, P.; Wudzinska, A.; Sun, X.; Andrade, J.; Nayak, S.; Kahler, D.J.; Badri, S.; LaCava, J.; Ueberheide, B.; Yun, C.Y.; et al. LINE-1 protein localization and functional dynamics during the cell cycle. Elife 2018, 7, e30058.
- Sato, Y.; Kinami, Y.; Hashiba, K.; Harashima, H. Different kinetics for the hepatic uptake of lipid nanoparticles between the apolipoprotein E/low density lipoprotein receptor and the N-acetyl-d-galactosamine/asialoglycoprotein receptor pathway. J. Control. Release 2020, 322, 217–226.
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Guler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289.
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, O.; Thompson, J.; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017, 25, 1316–1327.
- Bril, F.; Al Diffalha, S.; Dean, M.; Fettig, D.M. Autoimmune hepatitis developing after coronavirus disease 2019 (COVID-19) vaccine: Causality or casualty? J. Hepatol. 2021, 75, 222–224.
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370.
- Coffin, J.M.; Fan, H. The Discovery of Reverse Transcriptase. Annu. Rev. Virol. 2016, 3, 29–51.
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921.
- Ostertag, E.M.; Goodier, J.L.; Zhang, Y.; Kazazian, H.H., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003, 73, 1444–1451.
- Hancks, D.C.; Kazazian, H.H., Jr. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203.
- Jones, R.B.; Song, H.; Xu, Y.; Garrison, K.E.; Buzdin, A.A.; Anwar, N.; Hunter, D.V.; Mujib, S.; Mihajlovic, V.; Martin, E.; et al. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J. Virol. 2013, 87, 13307–13320.
- Macchietto, M.G.; Langlois, R.A.; Shen, S.S. Virus-induced transposable element expression up-regulation in human and mouse host cells. Life Sci. Alliance 2020, 3, e201900536.
- Yin, Y.; Liu, X.Z.; He, X.; Zhou, L.Q. Exogenous Coronavirus Interacts With Endogenous Retrotransposon in Human Cells. Front. Cell Infect. Microbiol. 2021, 11, 609160.
- Belancio, V.P.; Roy-Engel, A.M.; Deininger, P. The impact of multiple splice sites in human L1 elements. Gene 2008, 411, 38–45.
- Dai, L.; Taylor, M.S.; O’Donnell, K.A.; Boeke, J.D. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol. Cell Biol. 2012, 32, 4323–4336.
- Servant, G.; Streva, V.A.; Derbes, R.S.; Wijetunge, M.I.; Neeland, M.; White, T.B.; Belancio, V.P.; Roy-Engel, A.M.; Deininger, P.L. The Nucleotide Excision Repair Pathway Limits L1 Retrotransposition. Genetics 2017, 205, 139–153.
- Guo, H.; Chitiprolu, M.; Gagnon, D.; Meng, L.; Perez-Iratxeta, C.; Lagace, D.; Gibbings, D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat. Commun. 2014, 5, 5276.
- Xie, Y.; Mates, L.; Ivics, Z.; Izsvak, Z.; Martin, S.L.; An, W. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob. DNA 2013, 4, 10.
- Shi, X.; Seluanov, A.; Gorbunova, V. Cell divisions are required for L1 retrotransposition. Mol. Cell Biol. 2007, 27, 1264–1270.
- Goff, S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol 2007, 5, 253–263.
- Suzuki, Y.; Craigie, R. The road to chromatin—Nuclear entry of retroviruses. Nat. Rev. Microbiol. 2007, 5, 187–196.
- Shi, J.; Wang, X.; Lyu, L.; Jiang, H.; Zhu, H.J. Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: Focusing on drug-metabolizing enzymes. Drug Metab. Pharmacokinet. 2018, 33, 133–140.
- Kubo, S.; Seleme, M.C.; Soifer, H.S.; Perez, J.L.; Moran, J.V.; Kazazian, H.H., Jr.; Kasahara, N. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. USA 2006, 103, 8036–8041.
- Macia, A.; Widmann, T.J.; Heras, S.R.; Ayllon, V.; Sanchez, L.; Benkaddour-Boumzaouad, M.; Munoz-Lopez, M.; Rubio, A.; Amador-Cubero, S.; Blanco-Jimenez, E.; et al. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 2017, 27, 335–348.
The publisher is committed to upholding the integrity of the academic record and ensuring the accuracy of all published content. We strive to ensure that all published content is free from bias and inaccuracy, and we take any claims of inaccuracy or bias seriously. If you have any concerns about the accuracy or bias of a published article, please contact us at [email protected].
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of MDPI.
You may also like



