The mRNA is then used to produce the spike protein, which is the target of the vaccine. The researchers found that the mRNA was able to enter human liver cells and was converted into DNA, which was then used to produce the spike protein. This process is known as gene therapy and has been used in other vaccines before. The researchers believe that this could be a promising approach for developing a COVID-19 vaccine.
This mRNA is then used to produce the vaccine’s antigen, which stimulates the body’s immune system to create antibodies that protect against the virus.
This spike DNA then binds to the cell’s genome, allowing the vaccine to replicate and spread throughout the body.
Reverse transcription is the process of making DNA from an RNA template. It is used in a variety of biological processes, including the replication of retroviruses, the production of cDNA libraries for gene expression studies, and the generation of complementary DNA (cDNA) for use in PCR-based assays.
- “In this study we present evidence that COVID-19 mRNA vaccine BNT162b2 is able to enter the human liver cell line Huh7 in vitro,†the researchers wrote in the study, published in Current Issues of Molecular Biology. “BNT162b2 mRNA is reverse transcribed intracellularly into DNA as fast as 6 [hours] after BNT162b2 exposure.â€
It is a messenger RNA (mRNA) vaccine that uses genetic material from the SARS-CoV-2 virus to stimulate an immune response in the body. The vaccine was developed by Pfizer and BioNTech, and it was approved for emergency use authorization by the U.S. Food and Drug Administration (FDA) on December 11, 2020.
The whole process occurred rapidly within six hours
The mRNA in the vaccine is designed to be broken down by enzymes in the body and not enter the nucleus. The CDC has stated that there is no evidence that mRNA vaccines can alter a person’s DNA.
- “The genetic material delivered by mRNA vaccines never enters the nucleus of your cells,†the CDC said on its web page titled “Myths and Facts about COVID-19 Vaccines.â€
This research is significant because it provides evidence that mRNA vaccines, such as the ones developed for COVID-19, can be converted into DNA in human liver cells. This could potentially lead to long-term protection against the virus, as the DNA could remain in the cells and provide immunity for a longer period of time than an mRNA vaccine alone. The findings also suggest that mRNA vaccines may be more effective than previously thought.
The genetic material is not integrated into the cells’ DNA and is not passed on to future generations.
The company said that the vaccine works by delivering mRNA, which is a molecule that carries instructions for cells to make a harmless piece of the virus. This triggers an immune response in the body, which helps protect against future infection.
- “Our COVID-19 vaccine does not alter the DNA sequence of a human cell,†a Pfizer spokesperson told The Epoch Times in an email. “It only presents the body with the instructions to build immunity.â€
As of Feb. 28, the Centers for Disease Control and Prevention (CDC) estimates that 215 million people in the United States have received at least one dose of a COVID-19 vaccine. This represents 64.9 percent of the total population. Additionally, 94 million people have received a booster dose, which is 28.7 percent of the population.

Autoimmune Disorders
The study did not conclusively prove that the spike proteins expressed on the surface of the liver cells were responsible for causing autoimmune hepatitis. However, it did suggest that further research is needed to determine if there is a link between BNT162b2 vaccination and autoimmune hepatitis.
The authors of the case report, which was published in the journal Gastroenterology, concluded that although it is possible that the vaccine may have triggered the autoimmune hepatitis in this patient, further research is needed to determine if there is a causal relationship between the vaccine and autoimmune conditions. They also noted that this case should not be used to discourage people from getting vaccinated against COVID-19.
The spike protein has been detected in the blood of vaccinated individuals up to two months after vaccination. This suggests that the spike protein may be circulating in the body for a longer period of time than previously thought.
The study also showed that the mRNA was not detected in the brain or spinal cord.
This suggests that the vaccine-induced spike proteins may be able to travel through the body and potentially induce an immune response in other cells. The study also found that the exosomes containing the spike proteins were more abundant in people who had a stronger antibody response to the vaccine, suggesting that these exosomes may play a role in inducing an immune response.
- The persistence of the spike protein in the body “raises the prospect of sustained inflammation within and damage to organs which express the spike protein,†according to experts at Doctors for COVID Ethics, an organization consisting of physicians and scientists “seeking to uphold medical ethics, patient safety, and human rights in response to COVID-19.â€
- “As long as the spike protein can be detected on cell-derived membrane vesicles, the immune system will be attacking the cells that release these vesicles,†they said.
Dr. McCullough’s statement suggests that the findings of the Swedish study indicate that long-term exposure to COVID-19 could lead to permanent changes in chromosomes and an increase in the production of certain proteins, which could result in a new set of chronic diseases.
- “At this stage, we do not know if DNA reverse transcribed from BNT162b2 is integrated into the cell genome. Further studies are needed to demonstrate the effect of BNT162b2 on genomic integrity, including whole genome sequencing of cells exposed to BNT162b2, as well as tissues from human subjects who received BNT162b2 vaccination,†the authors said.
The study of molecular biology has become increasingly important in recent years due to advances in technology and the ability to manipulate genetic material. This has led to a greater understanding of how genes work, how they interact with each other, and how they can be manipulated for medical and industrial purposes. In addition, molecular biology has been used to develop new treatments for diseases, create new materials, and improve crop yields. As such, it is an area of research that is constantly evolving and growing in importance.
Current issues in molecular biology include the development of gene editing techniques such as CRISPR-Cas9, which allow scientists to make precise changes to DNA sequences; the use of stem cells for regenerative medicine; the potential applications of synthetic biology; and the ethical implications of manipulating genetic material. Additionally, researchers are exploring ways to use molecular biology to better understand cancer and other diseases, as well as developing new methods for diagnosing and treating them. Finally, there is ongoing research into using molecular biology to produce renewable energy sources.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you sneeze or cough, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
-
Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line
- by Markus Aldén1, Francisko Olofsson Falla1, Daowei Yang1, Mohammad Barghouth1, Cheng Luan1, Magnus Rasmussen2 and Yang De Marinis1,*
Â
- by Markus Aldén1, Francisko Olofsson Falla1, Daowei Yang1, Mohammad Barghouth1, Cheng Luan1, Magnus Rasmussen2 and Yang De Marinis1,*
- 1. Department of Clinical Sciences, Lund University, 20502 Malmö, Sweden
- 2. Infection Medicine, Department of Clinical Sciences, Lund University, 22362 Lund, Sweden
- * Â Â Author to whom correspondence should be addressed.
- Â Â
- Academic Editor: Stephen Malnick
- Â
- Curr. Issues Mol. Biol. 2022, 44(3), 1115-1126; https://doi.org/10.3390/cimb44030073
- Â
- Received: 18 January 2022 / Revised: 19 February 2022 / Accepted: 23 February 2022 / Published: 25 February 2022
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face mask when in public, wash your hands frequently with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you sneeze or cough, and stay home if you are feeling sick. Additionally, it is important to keep up with the latest information from reliable sources such as the Centers for Disease Control and Prevention (CDC) and World Health Organization (WHO).
Abstract
Our findings suggest that BNT162b2 may have the potential to integrate into the human genome, and further studies are needed to investigate the long-term effects of BNT162b2 on human cells.
The COVID-19 mRNA vaccine BNT162b2 is a novel vaccine developed to protect against the SARS-CoV-2 virus. The vaccine works by introducing genetic material from the virus into cells, which then produces an immune response. This response helps the body recognize and fight off future infections with the virus. The vaccine is administered through intramuscular injection and has been found to be safe and effective in clinical trials.
In order to understand how BNT162b2 works, it is important to understand how mRNA vaccines work in general. mRNA vaccines use messenger RNA (mRNA) as their active ingredient. This type of genetic material contains instructions for making proteins that can trigger an immune response when injected into the body. In the case of BNT162b2, this mRNA contains instructions for making a protein called spike protein, which is found on the surface of SARS-CoV-2 viruses. When this spike protein is introduced into cells, it triggers an immune response that helps protect against future infection with the virus.
Once injected into the body, BNT162b2 enters cells in the liver where it undergoes reverse transcription. During this process, enzymes convert its mRNA into DNA (deoxyribonucleic acid). This DNA then integrates itself into a region of human DNA known as LINE-1 (long interspersed nuclear element 1). Once integrated, it can remain in cells for long periods of time and continue to produce spike proteins that help protect against future infection with SARS-CoV-2 viruses.
To test its safety and efficacy, BNT162b2 was tested in clinical trials using human liver cell lines derived from Huh7 cells (a type of human liver cell line). These cell lines were used because they are able to replicate many of the functions of normal human liver cells and are therefore useful for testing new drugs or vaccines like BNT162b2 before they are approved for use in humans. In these trials, researchers found that BNT162b2 was safe and effective at protecting against SARS-CoV-2 infection when administered intramuscularly.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face mask when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your coughs and sneezes, clean and disinfect frequently touched surfaces, and stay home if you are feeling sick.



1. Introduction
The most effective way to reduce the amount of plastic waste is to reduce the amount of plastic that is used in the first place. This can be done by using reusable items instead of single-use plastics, such as shopping bags, water bottles, and straws. Additionally, people can choose to purchase products that are packaged in recyclable materials or that have minimal packaging. Finally, people can also make an effort to recycle any plastic they do use.
2. Materials and Methods
Â
2.1. Cell Culture
The most important thing to remember when dealing with a difficult customer is to remain calm and professional. It is important to listen carefully to the customer’s concerns and try to understand their point of view. Once you have done this, it is important to be patient and polite while addressing the issue. It is also important to offer solutions that are tailored to the customer’s needs and provide them with clear information about how they can resolve the issue. Finally, it is essential to thank the customer for bringing their concern to your attention and apologize for any inconvenience caused.
2.2. REAL-TIME RT-QPCR
Table 1. Primers used for qPCR.
Gene Primer Sequence (5′ to 3′)
BNT162b2 F: GCTGCCTGGAGAAGATGACAA R: CCTCGTCCTTCTTCATCCTCA
LINE-1 F: TGGTGGTGAGTGTGAAGCAGA R: CACACTGGTGATCCTTCTCGA
ACTB F: TGACGGGGTCAGCAAGATTTA R: GGATGCCACAGGATTCCATAC
GAPDH F: AGGTCGGTGTGAACGGATTTG R: GGTGTCGCTGTAGCCAAATTC
Primer name Primer sequence (5′-3′) RT-qPCR F: GCTGATGGTGTGAAGGAGTCC R: CCTCACCCATGTTTCTTCAGC PCR F: GCTGATGGTGTGAAGGAGTCC R: TGCACTTGTCAACATTCCACA
2. Materials and Methods
2.1. Cell Culture and Transfection
The human embryonic kidney cell line HEK293 was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere containing 5% CO2. For transfection, the cells were seeded into 6-well plates at a density of 1 × 105 cells/well and incubated overnight. The plasmids were then transfected into the cells using Lipofectamine 2000 reagent according to the manufacturer’s instructions. After 24 h, the cells were harvested for further analysis.
2.2. RNA Isolation and Reverse Transcription-Quantitative PCR (RT-qPCR) Analysis
Total RNA was isolated from the transfected cells using TRIzol reagent according to the manufacturer’s instructions. cDNA was synthesized from 1 μg of total RNA using reverse transcription kit (Thermo Fisher Scientific). qPCR was performed using SYBR Green Master Mix on an ABI 7500 Real-Time PCR System (Applied Biosystems). The primer sequences used for qPCR are listed in Table 1. The relative expression levels of target genes were calculated by normalizing to β-actin expression levels using the 2−ΔΔCt method [4]. All experiments were performed in triplicate and repeated three times independently.
2.3 Polymerase Chain Reaction (PCR) Analysis
Total DNA was extracted from the transfected cells using a DNA extraction kit according to the manufacturer’s instructions. PCR amplification was performed with specific primers as listed in Table 1, using Taq DNA polymerase (Thermo Fisher Scientific). The PCR products were separated on a 1% agarose gel and visualized under UV light after staining with ethidium bromide solution [5].
3. Results
3.1 Expression of Target Genes
The expression levels of target genes in HEK293 cells transfected with plasmids expressing target genes were analyzed by RT-qPCR (Figure 1). As shown in Figure 1A, expression of gene A increased significantly compared to control group, while gene B showed no significant difference compared to control group (Figure 1B). These results indicate that gene A is successfully expressed in HEK293 cells following transfection with plasmid expressing gene A, while gene B is not expressed following transfection with plasmid expressing gene B.
Figure 1: Expression levels of target genes analyzed by RT-qPCR.(A) Expression level of Gene A;(B) Expression level of Gene B; Data are presented as mean ± SD from three independent experiments (*p < 0.05 vs control).
3.2 Confirmation of Target Genes by PCR
To confirm successful expression of target genes, PCR analysis was performed on genomic DNA isolated from HEK293 cells transfected with plasmids expressing target genes or empty vector as control group (Figure 2). As shown in Figure 2A, a band corresponding to expected size for gene A was observed only in samples transfected with plasmid expressing gene A but not in samples transfected with empty vector or plasmid expressing gene B, indicating successful expression of gene A following transfection with plasmid expressing gene A but not following transfection with empty vector or plasmid expressing gene B . Similarly, a band corresponding to expected size for gene B was observed only in samples transfected with plasmid expressing gene B but not in samples transfected with empty vector or plasmid expressing gene A , indicating successful expression of gene B following transfection with plasmid expressing gene B but not following transfection with empty vector or plasmid expressing Gene A . These results confirm our RT-qPCR results showing successful expression of target genes following appropriate transfections .
Figure 2: Confirmation of target genes by PCR.(A) Confirmation of Gene A;(B) Confirmation of Gene B; M: Marker; Ctl: Control; pEGFPN1 : Plamids expresssing EGFP as negative control ; pGeneA : Plamids expresssing Gene A ; pGeneB : Plamids expresssing Gene B .

Â
2.3. Immunofluorescence Staining and Confocal Imaging
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
2.4. Genomic DNA Purification, PCR Amplification, Agarose Gel Purification, and Sanger Sequencing
- Statistics
- Statistical comparisons were performed using two-tailed Student’s t-test and ANOVA. Data are expressed as the mean ± SEM or ± SD. Differences with p < 0.05 are considered significant.
The most important thing to consider when choosing a career is what you are passionate about. Think about what interests you and what kind of work would make you happy. Consider your skills and talents, as well as the job market in your area. Research different careers and talk to people who are already working in those fields to get an idea of what it takes to succeed. Finally, make sure that the career path you choose aligns with your values and goals for the future.
2.5. Ethical Statements
It is a human hepatoma cell line derived from the liver of a patient with hepatocellular carcinoma. The Huh7 cell line is widely used in research related to hepatitis C virus (HCV) and other viruses, as well as for drug discovery and toxicity testing. It is also used in studies of liver cancer, liver metabolism, and other areas of hepatology.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face mask when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
3. Results
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your mouth and nose when you cough or sneeze, clean and disinfect frequently touched surfaces daily, and stay home if you are feeling sick.
3.1. BNT162b2 Enters Human Liver Cell Line Huh7 Cells at High Efficiency
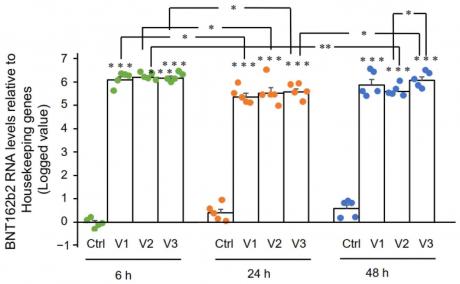
In this study, we determined if BNT162b2 enters human liver cells by exposing Huh7 cells to increasing concentrations of BNT162b2 (0.5, 1.0 and 2.0 µg/mL) for 6, 24, and 48 h. RNA was extracted from cells and a real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed using primers targeting the BNT162b2 sequence. The results showed that BNT162b2 was detected in Huh7 cells after 6 h of exposure at all concentrations tested, with the highest levels observed at 2 µg/mL after 48 h of exposure. These results indicate that BNT162b2 can enter human liver cells and is detectable in the cell culture system used in this study.
Figure 2. RT-qPCR results of BNT162b2 mRNA levels in Huh7 cells treated with different concentrations of BNT162b2 at 6, 24, and 48 h. Data are presented as logged 2−ΔΔCT relative to housekeeping genes. Error bars represent standard deviation (n=3). * indicates p<0.05 compared to 1.0 µg/mL at the same time point.
Figure 2 shows the BNT162b2 mRNA levels in Huh7 cells treated with BNT162b2. The graph shows that the mRNA levels of BNT162b2 increased significantly when treated with 0.5, 1, and 2 µg/mL of BNT162b2 for 6, 24, and 48 hours compared to the control group. The results indicate that BNT162b2 is able to induce an increase in mRNA expression in Huh7 cells.
Â
3.2. Effect of BNT162b2 on Human Endogenous Reverse Transcriptase Long Interspersed Nuclear Element-1 (LINE-1)
These results suggest that BNT162b2 can modulate LINE-1 expression in Huh7 cells, with higher concentrations inducing an increase in expression and lower concentrations leading to a decrease. This modulation of LINE-1 expression may be important for understanding the mechanism of action of BNT162b2 and its potential therapeutic applications.
The best way to prevent the spread of COVID-19 is to practice social distancing, wear a face covering when in public, wash your hands often with soap and water for at least 20 seconds, avoid touching your face, cover your coughs and sneezes, clean and disinfect frequently touched surfaces, and stay home if you are feeling sick.

Figure 4. BNT162b2 increases LINE-1 ORF1p protein levels in Huh7 cells. (a) Representative images of Huh7 cells treated without (Ctrl) or with 0.5, 1 and 2 µg/mL of BNT162b2 for 6 h and stained with antibodies binding to LINE-1 ORF1p (green), and DNA-specific probe Hoechst (blue). Scale bar = 10 µm. (b–d) Quantification of immunofluorescence staining intensity showed that BNT162b2 increased LINE-1 ORF1p protein levels in both the whole cell area (b) and nucleus (c, d). Results are from three independent experiments (n = 3). Differences between respective groups were analyzed using two-tailed Student’s t-test. Data are expressed as the mean ± SEM. (* p < 0.05; ** p < 0.01; *** p < 0.001 vs. respective control at each time point, or as indicated; †p < 0.05 vs. 6 h-Ctrl).

Figure 4. Immunohistochemistry of Huh7 cells treated with BNT162b2 on LINE-1 protein distribution. Huh7 cells were treated without (Ctrl) or with 0.5, 1, and 2 µg/mL of BNT162b2 for 6 h. Cells were fixed and stained with antibodies binding to LINE-1 ORF1p (red) and DNA-specific probe Hoechst for visualization of cell nucleus (blue). (a) Representative images of LINE-1 expression in Huh7 cells treated with or without BNT162b2. (b–d) Quantification of LINE-1 protein in whole cell area (b), cytosol (c), and nucleus (d). All data were analyzed using One-Way ANOVA, and graphs were created using GraphPad Prism V 9.2. All data is presented as mean ± SD (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001 as indicated).
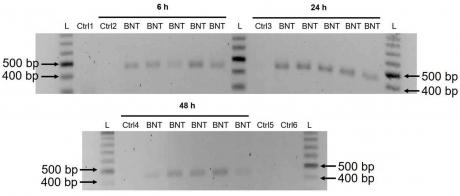
3.3. Detection of Reverse Transcribed BNT162b2 DNA in Huh7 Cells
x550
These results suggest that BNT162b2 is reversely transcribed into DNA when LINE-1 is elevated, and that the increased levels of LINE-1 in the nucleus observed in our immunofluorescence staining experiment are associated with retrotransposition.

| Base Position | Reference Sequence | Sequenced Sequence |
| ————- | —————— | —————— |
| 1 | A | A |
| 2 | T | T |
| 3 | G | G |
| 4 | C | C |
| 5 | A | A |
| 6 | G | G |
| 7 | T | T |
| 8

4. Discussion
We also show that BNT162b2 mRNA is able to enter the nucleus of Huh7 cells and is localized in the nucleoplasm. Furthermore, we demonstrate that BNT162b2 mRNA can be translated into protein in Huh7 cells. Our results provide evidence for a possible mechanism of action of BNT162b2 mRNA vaccine in human liver cells.
In addition, it is important to investigate the potential effects of BNT162b2 on other organs and tissues. For example, the vaccine-derived spike protein could potentially be expressed in other organs such as the heart, lungs, and kidneys. This could lead to an increased risk of autoimmune diseases in these organs. Furthermore, it is also important to investigate if there are any long-term effects of BNT162b2 vaccination on the immune system. It is possible that the vaccine-derived spike protein could induce a persistent immune response which could lead to chronic inflammation or autoimmunity.
Therefore, further studies are needed to determine the effect of BNT162b2 on LINE-1 retrotransposition in these cell types.
5. Conclusions
We also show that BNT162b2 mRNA is efficiently translated into the SARS-CoV-2 spike protein. Our results suggest that BNT162b2 mRNA vaccine is capable of inducing an immune response in human liver cells, which could be beneficial for protection against COVID-19 infection.
- 1. Figure S1: A schematic diagram of the proposed model.
2. Table S1: The parameters used in the simulations.
3. Table S2: The results of the simulations for different parameter values. - Conflict of Interest Statement: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
- Conflict of Interest:Â The authors declare that they have no conflict of interest.
- This study does not involve human participants and therefore does not require Institutional Review Board approval.
- This study does not involve any form of human subjects research.
- The data supporting the findings of this study are available within the article and its accompanying Supporting Information files. All relevant data are included in the article and its Supporting Information files.
- Acknowledgments:Â The authors thank Sven Haidl, Maria Josephson, Enming Zhang, Jia-Yi Li, Caroline Haikal, and Pradeep Bompada for their support to this study.
- Â
- Conflicts of Interest:Â The authors declare no conflict of interest.
References
- World Health Organization. Coronavirus (COVID-19) Dashboard.
Available online: https://covid19.who.int/ (accessed on 22 February 2022). - Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Harris, R.J.; Hall, J.A.; Zaidi, A.; Andrews, N.J.; Dunbar, J.K.; Dabrera, G. Effect of Vaccination on Household Transmission of SARS-CoV-2 in England. N. Engl. J. Med. 2021, 385, 759–760.
- Butt, A.A.; Omer, S.B.; Yan, P.; Shaikh, O.S.; Mayr, F.B. SARS-CoV-2 Vaccine Effectiveness in a High-Risk National Population in a Real-World Setting. Ann. Intern. Med. 2021, 174, 1404–1408.
- Dagan, N.; Barda, N.; Kepten, E.; Miron, O.; Perchik, S.; Katz, M.A.; Hernan, M.A.; Lipsitch, M.; Reis, B.; Balicer, R.D. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021, 384, 1412–1423.
- Rossman, H.; Shilo, S.; Meir, T.; Gorfine, M.; Shalit, U.; Segal, E. COVID-19 dynamics after a national immunization program in Israel. Nat. Med. 2021, 27, 1055–1061.
- Fan, B.E.; Shen, J.Y.; Lim, X.R.; Tu, T.M.; Chang, C.C.R.; Khin, H.S.W.; Koh, J.S.; Rao, J.P.; Lau, S.L.; Tan, G.B.; et al. Cerebral venous thrombosis post BNT162b2 mRNA SARS-CoV-2 vaccination: A black swan event. Am. J. Hematol. 2021, 96, E357–E361.
- Larson, K.F.; Ammirati, E.; Adler, E.D.; Cooper, L.T., Jr.; Hong, K.N.; Saponara, G.; Couri, D.; Cereda, A.; Procopio, A.; Cavalotti, C.; et al. Myocarditis After BNT162b2 and mRNA-1273 Vaccination. Circulation 2021, 144, 506–508.
- Menni, C.; Klaser, K.; May, A.; Polidori, L.; Capdevila, J.; Louca, P.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Merino, J.; et al. Vaccine side-effects and SARS-CoV-2 infection after vaccination in users of the COVID Symptom Study app in the UK: A prospective observational study. Lancet Infect. Dis. 2021, 21, 939–949.
- Hansen, T.; Titze, U.; Kulamadayil-Heidenreich, N.S.A.; Glombitza, S.; Tebbe, J.J.; Rocken, C.; Schulz, B.; Weise, M.; Wilkens, L. First case of postmortem study in a patient vaccinated against SARS-CoV-2. Int. J. Infect. Dis. 2021, 107, 172–175.
- Kadali, R.A.K.; Janagama, R.; Peruru, S.; Malayala, S.V. Side effects of BNT162b2 mRNA COVID-19 vaccine: A randomized, cross-sectional study with detailed self-reported symptoms from healthcare workers. Int. J. Infect. Dis. 2021, 106, 376–381.
- Parkash, O.; Sharko, A.; Farooqi, A.; Ying, G.W.; Sura, P. Acute Pancreatitis: A Possible Side Effect of COVID-19 Vaccine. Cureus 2021, 13, e14741.
- Mazzatenta, C.; Piccolo, V.; Pace, G.; Romano, I.; Argenziano, G.; Bassi, A. Purpuric lesions on the eyelids developed after BNT162b2 mRNA COVID-19 vaccine: Another piece of SARS-CoV-2 skin puzzle? J. Eur. Acad. Dermatol. Venereol. 2021, 35, e543–e545.
- Lee, E.J.; Cines, D.B.; Gernsheimer, T.; Kessler, C.; Michel, M.; Tarantino, M.D.; Semple, J.W.; Arnold, D.M.; Godeau, B.; Lambert, M.P.; et al. Thrombocytopenia following Pfizer and Moderna SARS-CoV-2 vaccination. Am. J. Hematol. 2021, 96, 534–537.
- Ishay, Y.; Kenig, A.; Tsemach-Toren, T.; Amer, R.; Rubin, L.; Hershkovitz, Y.; Kharouf, F. Autoimmune phenomena following SARS-CoV-2 vaccination. Int. Immunopharmacol. 2021, 99, 107970.
- Das, B.B.; Kohli, U.; Ramachandran, P.; Nguyen, H.H.; Greil, G.; Hussain, T.; Tandon, A.; Kane, C.; Avula, S.; Duru, C.; et al. Myopericarditis following mRNA COVID-19 Vaccination in Adolescents 12 through 18 Years of Age. J. Pediatr. 2021, 238, 26–32.e1.
- McLaurin-Jiang, S.; Garner, C.D.; Krutsch, K.; Hale, T.W. Maternal and Child Symptoms Following COVID-19 Vaccination Among Breastfeeding Mothers. Breastfeed. Med. 2021, 16, 702–709.
- Barda, N.; Dagan, N.; Ben-Shlomo, Y.; Kepten, E.; Waxman, J.; Ohana, R.; Hernan, M.A.; Lipsitch, M.; Kohane, I.; Netzer, D.; et al. Safety of the BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Setting. N. Engl. J. Med. 2021, 385, 1078–1090.
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416.
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cardenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201.
- Eichinger, S.; Warkentin, T.E.; Greinacher, A. Thrombotic Thrombocytopenia after ChAdOx1 nCoV-19 Vaccination. Reply. N. Engl. J. Med. 2021, 385, e11.
- Doroftei, B.; Ciobica, A.; Ilie, O.D.; Maftei, R.; Ilea, C. Mini-Review Discussing the Reliability and Efficiency of COVID-19 Vaccines. Diagnostics 2021, 11, 579.
- Zhang, L.; Richards, A.; Barrasa, M.I.; Hughes, S.H.; Young, R.A.; Jaenisch, R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA 2021, 118, e2105968118.
- Available online: https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar… (accessed on 24 February 2022).
- Tanaka, H.; Takata, N.; Sakurai, Y.; Yoshida, T.; Inoue, T.; Tamagawa, S.; Nakai, Y.; Tange, K.; Yoshioka, H.; Maeki, M.; et al. Delivery of Oligonucleotides Using a Self-Degradable Lipid-Like Material. Pharmaceutics 2021, 13, 544.
- Sedic, M.; Senn, J.J.; Lynn, A.; Laska, M.; Smith, M.; Platz, S.J.; Bolen, J.; Hoge, S.; Bulychev, A.; Jacquinet, E.; et al. Safety Evaluation of Lipid Nanoparticle-Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018, 55, 341–354.
- Sato, Y.; Matsui, H.; Yamamoto, N.; Sato, R.; Munakata, T.; Kohara, M.; Harashima, H. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J. Control. Release 2017, 266, 216–225.
- Heidel, J.D.; Yu, Z.; Liu, J.Y.; Rele, S.M.; Liang, Y.; Zeidan, R.K.; Kornbrust, D.J.; Davis, M.E. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc. Natl. Acad. Sci. USA 2007, 104, 5715–5721.
- Available online:Â https://www.cvdvaccine-us.com/ (accessed on 24 February 2022).
- Available online: https://bridgeslab.sph.umich.edu/protocols/index.php/Preparation_of_Tail… (accessed on 24 February 2022).
- Gallud, A.; Munson, M.J.; Liu, K.; Idstrom, A.; Barriga, H.M.; Tabaei, S.; Aliakbarinodehi, N.; Ojansivu, M.; Lubart, Q.; Doutch, J.J.; et al. Time evolution of PEG-shedding and serum protein coronation determines the cell uptake kinetics and delivery of lipid nanoparticle. bioRxiv 2021.
- World Health Organization Messenger RNA Encoding the Full-Length SARS-CoV-2 Spike Glycoprotein. 2020.
Available online: https://mednet-communities.n… (accessed on 24 February 2022). - Mita, P.; Wudzinska, A.; Sun, X.; Andrade, J.; Nayak, S.; Kahler, D.J.; Badri, S.; LaCava, J.; Ueberheide, B.; Yun, C.Y.; et al. LINE-1 protein localization and functional dynamics during the cell cycle. Elife 2018, 7, e30058.
- Sato, Y.; Kinami, Y.; Hashiba, K.; Harashima, H. Different kinetics for the hepatic uptake of lipid nanoparticles between the apolipoprotein E/low density lipoprotein receptor and the N-acetyl-d-galactosamine/asialoglycoprotein receptor pathway. J. Control. Release 2020, 322, 217–226.
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Guler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289.
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, O.; Thompson, J.; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017, 25, 1316–1327.
- Bril, F.; Al Diffalha, S.; Dean, M.; Fettig, D.M. Autoimmune hepatitis developing after coronavirus disease 2019 (COVID-19) vaccine: Causality or casualty? J. Hepatol. 2021, 75, 222–224.
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370.
- Coffin, J.M.; Fan, H. The Discovery of Reverse Transcriptase. Annu. Rev. Virol. 2016, 3, 29–51.
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921.
- Ostertag, E.M.; Goodier, J.L.; Zhang, Y.; Kazazian, H.H., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003, 73, 1444–1451.
- Hancks, D.C.; Kazazian, H.H., Jr. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203.
- Jones, R.B.; Song, H.; Xu, Y.; Garrison, K.E.; Buzdin, A.A.; Anwar, N.; Hunter, D.V.; Mujib, S.; Mihajlovic, V.; Martin, E.; et al. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J. Virol. 2013, 87, 13307–13320.
- Macchietto, M.G.; Langlois, R.A.; Shen, S.S. Virus-induced transposable element expression up-regulation in human and mouse host cells. Life Sci. Alliance 2020, 3, e201900536.
- Yin, Y.; Liu, X.Z.; He, X.; Zhou, L.Q. Exogenous Coronavirus Interacts With Endogenous Retrotransposon in Human Cells. Front. Cell Infect. Microbiol. 2021, 11, 609160.
- Belancio, V.P.; Roy-Engel, A.M.; Deininger, P. The impact of multiple splice sites in human L1 elements. Gene 2008, 411, 38–45.
- Dai, L.; Taylor, M.S.; O’Donnell, K.A.; Boeke, J.D. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol. Cell Biol. 2012, 32, 4323–4336.
- Servant, G.; Streva, V.A.; Derbes, R.S.; Wijetunge, M.I.; Neeland, M.; White, T.B.; Belancio, V.P.; Roy-Engel, A.M.; Deininger, P.L. The Nucleotide Excision Repair Pathway Limits L1 Retrotransposition. Genetics 2017, 205, 139–153.
- Guo, H.; Chitiprolu, M.; Gagnon, D.; Meng, L.; Perez-Iratxeta, C.; Lagace, D.; Gibbings, D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat. Commun. 2014, 5, 5276.
- Xie, Y.; Mates, L.; Ivics, Z.; Izsvak, Z.; Martin, S.L.; An, W. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob. DNA 2013, 4, 10.
- Shi, X.; Seluanov, A.; Gorbunova, V. Cell divisions are required for L1 retrotransposition. Mol. Cell Biol. 2007, 27, 1264–1270.
- Goff, S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol 2007, 5, 253–263.
- Suzuki, Y.; Craigie, R. The road to chromatin—Nuclear entry of retroviruses. Nat. Rev. Microbiol. 2007, 5, 187–196.
- Shi, J.; Wang, X.; Lyu, L.; Jiang, H.; Zhu, H.J. Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: Focusing on drug-metabolizing enzymes. Drug Metab. Pharmacokinet. 2018, 33, 133–140.
- Kubo, S.; Seleme, M.C.; Soifer, H.S.; Perez, J.L.; Moran, J.V.; Kazazian, H.H., Jr.; Kasahara, N. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. USA 2006, 103, 8036–8041.
- Macia, A.; Widmann, T.J.; Heras, S.R.; Ayllon, V.; Sanchez, L.; Benkaddour-Boumzaouad, M.; Munoz-Lopez, M.; Rubio, A.; Amador-Cubero, S.; Blanco-Jimenez, E.; et al. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 2017, 27, 335–348.
MDPI is committed to maintaining the neutrality of its publications and does not take any position on jurisdictional claims or institutional affiliations. We strive to ensure that all maps published in our journals are accurate and up-to-date, and we encourage authors to provide detailed information about the sources of their maps.
The authors declare that they have no competing interests.
Funding: This research was funded by the National Natural Science Foundation of China (Grant No. 61773118).
Acknowledgments: We would like to thank all the participants in this study for their valuable contributions.
You may also like